Abstract
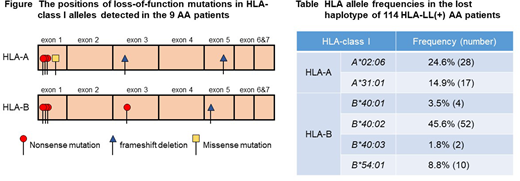
[Background] Acquired aplastic anemia (AA) is a rare syndrome characterized by pancytopenia and bone marrow hypoplasia. The cytotoxic T lymphocyte (CTL) attack against autologous hematopoietic stem progenitor cells (HSPCs) is thought to be responsible for bone marrow failure in the majority of AA cases; however, little is known about the target antigens of the CTLs. HLA class I-allele lacking leukocytes (HLA-LL) due to copy-number neutral loss of heterozygosity in the short arm of chromosome 6 (6pLOH) or somatic loss-of-function mutations in HLA class I genes are detected in approximately 20% of patients with newly diagnosed AA, and the presence of HLA-LL represents compelling evidence to support that CTLs specific to HSPCs are involved in the development of AA. Our recent studies using single nucleotide polymorphism array (SNP-A) genotyping and droplet digital polymerase chain reaction (ddPCR) revealed that HLA-B*40:02 is the most frequently lost among all class I alleles that are lost as a result of 6pLOH (Zaimoku, et al. Blood 2017). Various somatic loss-of-function mutations in B*40:02 revealed by deep sequencing in the study substantiated the important role of HLA-B4002 in the autoantigen presentation of AA. However, in the other 6pLOH(+) AA patients who did not possess HLA-B4002, which accounted for 20% of the total AA cases involving patients possessing HLA-LL, the allele in the missing haplotype that was responsible for the autoantigen presentation was largely unknown because the lost fragment of chromosome 6p usually contained 2 or more HLA class I alleles. [Objectives/Methods] To identify class I alleles other than HLA-B*40:02 that are critically involved in the auto-antigen presentation of AA, we screened a total of 624 patients for the presence of HLA-LL using monoclonal antibodies specific to class I HLA alleles, SNP-A, and ddPCR, and performed targeted deep sequencing of HLA class I genes by using SeqCap EZ Choice pobes (Roche) and MiSeq sequencer (Illumina). The paired fractions, including granulocytes that lacked an HLA-A allele and granulocytes that retained the HLA-A allele, as well as CD3+ T cells, were sorted using monoclonal antibodies specific to HLA-A alleles with a BD FACSAria Fusion system (BD Biosciences), and were subjected to DNA extraction. All DNA samples of granulocytes and control cells (CD3+ T cells or buccal mucosa cells) were prepared for targeted deep sequencing. [Results] One hundred and fourteen patients were found to be positive for HLA-LL and 62 (54.4%) of the 114 HLA-LL(+) patients did not carry B*40:02 (severe, n=30; non-severe, n=32; male, n=38; female, n=24; median age, 62 [range, 6-93] years). Apart from B*40:02 (45.6%), A*02:06 (24.6%) was the second-most frequent HLA class I allele in the lost haplotype. The targeted deep sequencing of 20 patients with HLA-LL revealed 6pLOH alone in 11 patients, and somatic loss-of-function mutations plus 6pLOH in 9 patients; none of the patients were positive for somatic loss-of-function mutations alone. Of note, somatic loss-of-function mutations were found in only 5 alleles (A*02:06 in four, B*40:01 in two, B*40:03, A*31:01, and B*54:01 in one each) out of 27 different alleles contained in the lost haplotype. Among the 9 patients with somatic loss-of-function mutations, the median number of mutations per patient was 1 (range, 1-2); these included a missense mutation (n=1), frameshift deletions (n=3) and nonsense mutations (n=7) (Figure). Four patients had a breakpoint of 6pLOH in between the HLA-A and C loci; their lost alleles were A*02:06 (n=2) and A*31:01 (n=2), and the occurrence of 6pLOH in the four patients was therefore attributed to the two HLA-A alleles. Sixty-six percent of the HLA-LL(+) B*40:02(-) patients had at least one of the five alleles in the lost haplotypes. The frequencies of each "high risk" allele found in patients possessing HLA-LL are summarized in Table. [Conclusions] In addition to B*40:02, five class I alleles including HLA-A*02:06, A*31:01, B*54:01, B*40:03 and B*40:01 are thought to play an essential role in the auto-antigen presentation by the HSPCs of Japanese AA patients. The frequencies of the six class I alleles in general Japanese population are much higher than those in the general Caucasian populations but similar to the frequencies in East Asian populations. The higher frequencies of the six alleles in comparison to Caucasian countries may account for the higher incidence of AA in East Asia.
Takamatsu:Ono: Research Funding; Bristol-Myers Squibb: Research Funding; Janssen: Honoraria; Celgene: Honoraria, Research Funding. Nakao:Novartis: Honoraria; Alexion Pharmaceuticals, Inc.: Consultancy, Honoraria; Kyowa Hakko Kirin Co., Ltd.: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal